Personas bien vacunadas pueden recaer infectadas con un virus que ha hecho mutaciones en su propio cuerpo.
Las variantes emergentes del síndrome respiratorio agudo severo coronavirus 2 (SARS-CoV-2) son motivo de preocupación clínica. En una cohorte de 417 personas que habían recibido la segunda dosis de la vacuna BNT162b2 (Pfizer – BioNTech) o mRNA-1273 (Moderna) al menos 2 semanas antes, identificamos a 2 mujeres con infección por avance de la vacuna.
A pesar de la evidencia de la eficacia de la vacuna en ambas mujeres, se desarrollaron síntomas de la enfermedad por coronavirus 2019 y dieron positivo al SARS-CoV-2 mediante la prueba de reacción en cadena de la polimerasa. La secuenciación viral reveló variantes de probable importancia clínica, como E484K en una mujer y tres mutaciones (T95I, del142-144 y D614G) en ambas.
Estas observaciones indican un riesgo potencial de enfermedad después de una vacunación exitosa y la posterior infección con una variante del virus. y brindan apoyo a los esfuerzos continuos para prevenir y diagnosticar infecciones y caracterizar variantes en personas vacunadas. (Financiado por los Institutos Nacionales de Salud y otros).
Tratamiento novedoso y efectivo para el Covid-19
Texto original en inglés, fuente: njem.org
Vaccine Breakthrough Infections with SARS-CoV-2 Variants
List of authors.
Summary
Emerging variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are of clinical concern. In a cohort of 417 persons who had received the second dose of BNT162b2 (Pfizer–BioNTech) or mRNA-1273 (Moderna) vaccine at least 2 weeks previously, we identified 2 women with vaccine breakthrough infection. Despite evidence of vaccine efficacy in both women, symptoms of coronavirus disease 2019 developed, and they tested positive for SARS-CoV-2 by polymerase-chain-reaction testing. Viral sequencing revealed variants of likely clinical importance, including E484K in 1 woman and three mutations (T95I, del142–144, and D614G) in both. These observations indicate a potential risk of illness after successful vaccination and subsequent infection with variant virus, and they provide support for continued efforts to prevent and diagnose infection and to characterize variants in vaccinated persons. (Funded by the National Institutes of Health and others.)
Methods
SPECIMEN COLLECTION AND PROCESSING
Beginning in the fall of 2020, all employees and students at the Rockefeller University campus (approximately 1400 persons) were tested at least weekly with a saliva-based PCR test developed in the Darnell Clinical Laboratory Improvement Amendments–Clinical Laboratory Evaluation Program laboratory (approval number, PFI-9216) and approved for clinical use by a New York State emergency use authorization. Protocols for the collection of saliva samples for clinical SARS-CoV-2 testing were reviewed by the institutional review board at Rockefeller University and were deemed not to be research involving human subjects. Institutional review board–approved written informed consent for the analysis of antibody titers was obtained from Patient 1, and the study was conducted in accordance with International Council for Harmonisation Good Clinical Practice guidelines.
In accordance with New York State regulations regarding eligibility, 417 employees who had received a second dose of either the BNT162b2 (Pfizer–BioNTech) or mRNA-1273 (Moderna) vaccine at least 2 weeks previously were tested between January 21 and March 17, 2021, and weekly testing continued thereafter. The demographic characteristics of these 417 persons and of 1491 unvaccinated persons tested in parallel at Rockefeller University during the same period are shown in Table S1 of the Supplementary Appendix, available with the full text of this article at NEJM.org.
The employees and students were instructed to provide a saliva sample in a medicine cup and transfer 300 μl into a vial containing 300 μl of Darnell Rockefeller University Laboratory (DRUL) buffer (5 M of guanidine thiocyanate, 0.5% sarkosyl, and 300 mM of sodium acetate [pH 5.5]).2 Samples were processed on the Thermo KingFisher Apex system for rapid RNA purification, and complementary DNA (cDNA) was amplified with the use of TaqPath 1-Step RT-qPCR (reverse-transcriptase quantitative PCR) Master Mix (Thermo Fisher Scientific) and multiplexed primers and probes that were validated under a Food and Drug Administration emergency use authorization (Table S2) with the 7500 Fast Dx Real-Time PCR detection system (Applied Biosystems). Samples were considered to be interpretable if the housekeeping control (RNase P) cycle threshold (Ct) was less than 40, and viral RNA was considered to be detected with both viral primers and probes (N1 and N2, detecting two regions of the nucleocapsid [N] gene of SARS-CoV-2) at a Ct of less than 40.
VIRAL LOAD CALCULATION
We calculated the viral load per milliliter of saliva using chemically inactivated SARS-CoV-2 (ZeptoMetrix) spiked into saliva at various dilutions. Extractions and RT-PCR were performed as described previously to determine the corresponding Ct values for each dilution (Fig. S1).
TARGETED SEQUENCING
Reverse transcription of RNA samples was performed with the iScript mix (Bio-Rad) according to the manufacturer’s instructions. PCR amplification of cDNA was performed with the use of two primer sets (primer set 1: forward primer 1 [CCAGATGATTTTACAGGCTGC] and reverse primer 1 [CTACTGATGTCTTGGTCATAGAC]; primer set 2: forward primer 2 [CTTGTTTTATTGCCACTAGTC] and reverse primer 1). PCR products were then extracted from gel and sent to Genewiz for Sanger sequencing.
NEUTRALIZATION ASSAY
Neutralization assays with pseudotyped replication defective human immunodeficiency virus type 1 modified with SARS-CoV-2 spike protein were performed as previously described.3 Mean serum neutralizing antibody titers (50% neutralization testing [NT50]) were calculated as an average of three independent experiments, each performed with the use of technical duplicates, and statistical significance was determined with the two-tailed Mann–Whitney test.
WHOLE VIRAL RNA GENOME SEQUENCING
Total RNA was extracted as described above, and a meta-transcriptomic library was constructed for paired-end (150-bp reads) sequencing with an Illumina MiSeq platform. Libraries were prepared with the SureSelect XT HS2 DNA System (Agilent Technologies) and Community Design Pan Human Coronavirus Panel (Agilent Technologies) according to the manufacturer’s instructions. FASTQ files (a text-based format for storing both a biologic sequence and its corresponding quality scores) were trimmed with Agilent Genomics NextGen Toolkit (AGeNT) software (version 2.0.5) and used for downstream analysis. The SARS-CoV-2 genome was assembled with MEGAHIT with default parameters, and the longest sequence (30,005 nucleotides) was analyzed with Nextclade software (https://clades.nextstrain.org/. opens in new tab) in order to assign the clade and call mutations. Detected mutations were confirmed by aligning RNA sequencing reads on the reference genome sequence of SARS-CoV-2 (GenBank number, NC_045512) with the Burrows–Wheeler Aligner (BWA-MEM).
PATIENT HISTORIES
Patient 1 was a healthy 51-year-old woman with no risk factors for severe Covid-19 who received the first dose of mRNA-1273 vaccine on January 21, 2021, and the second dose on February 19. She had adhered strictly to routine precautions. Ten hours after she received the second vaccine dose, flulike muscle aches developed. These symptoms resolved the following day. On March 10 (19 days after she received the second vaccine dose), a sore throat, congestion, and headache developed, and she tested positive for SARS-CoV-2 RNA at Rockefeller University later that day. On March 11, she lost her sense of smell. Her symptoms gradually resolved over a 1-week period.
Patient 2 was a healthy 65-year-old woman with no risk factors for severe Covid-19 who received the first dose of BNT162b2 vaccine on January 19 and the second dose on February 9. Pain that developed in the inoculated arm lasted for 2 days. On March 3, her unvaccinated partner tested positive for SARS-CoV-2, and on March 16, fatigue, sinus congestion, and a headache developed in Patient 2. On March 17, she felt worse and tested positive for SARS-CoV-2 RNA, 36 days after completing vaccination. Her symptoms plateaued and began to resolve on March 20.
Results
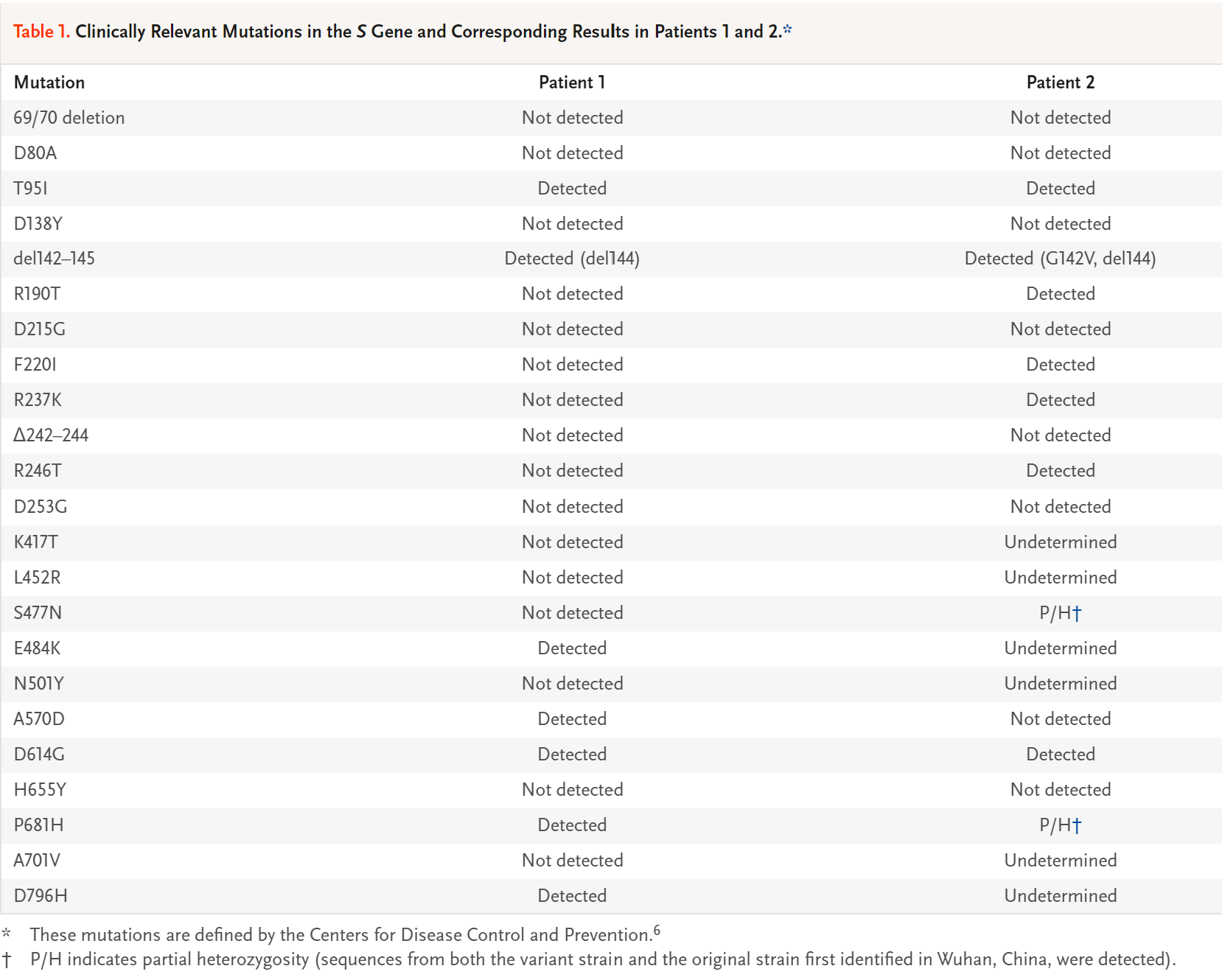
Clinically Relevant Mutations in the S Gene and Corresponding Results in Patients 1 and 2.
Serial PCR tests of saliva samples were performed before and during the course of the illness in both patients (Table S3). At the time of diagnosis, PCR testing showed a Ct value that corresponded to a viral load of approximately 195,000 copies per milliliter of saliva in Patient 1 and approximately 400 copies per milliliter in Patient 2 (Fig. S1). Given the unusual clinical histories of these two patients, RNA obtained from the saliva samples was sequenced after reverse transcription and targeted PCR amplification of the SARS-CoV-2 S gene. This test revealed a number of differences between the sequences and the original sequence first identified in Wuhan, China, including several variants that have been suggested to be of potential clinical concern. In Patient 1, these mutations included E484K (which confers resistance to a commonly elicited class of neutralizing antibodies4,5) and D614G, and in Patient 2, these mutations included D614G and S477N (Table 1).
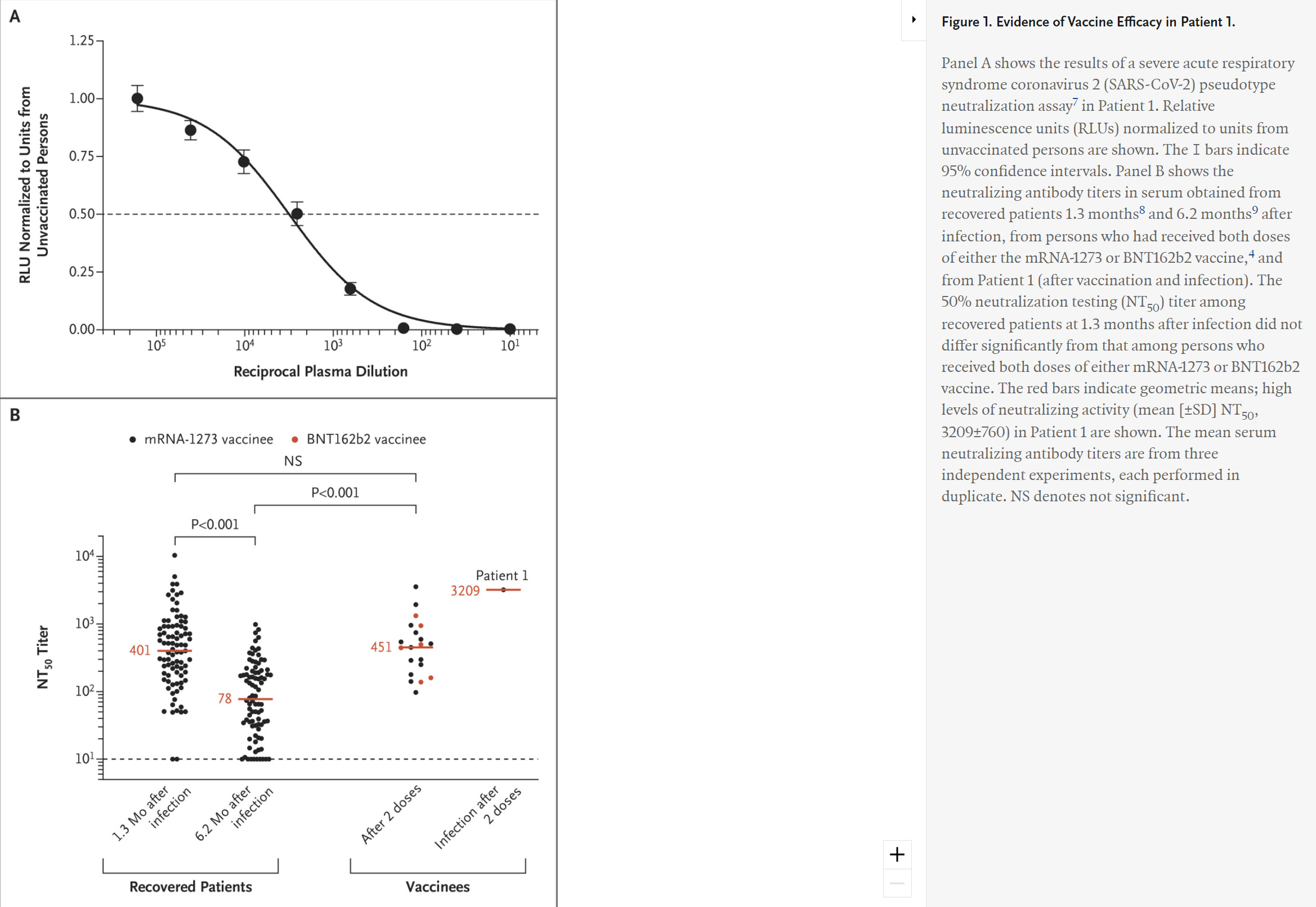
Evidence of Vaccine Efficacy in Patient 1.
A serum sample obtained 4 days after the onset of symptoms in Patient 1 was analyzed for neutralizing antibodies to SARS-CoV-2 in a pseudotype neutralization assay.3 These results revealed extremely high titers of neutralizing antibodies (Figure 1). These data, together with the clinical history, provide strong evidence that this patient probably had neutralizing antibodies elicited by vaccination.
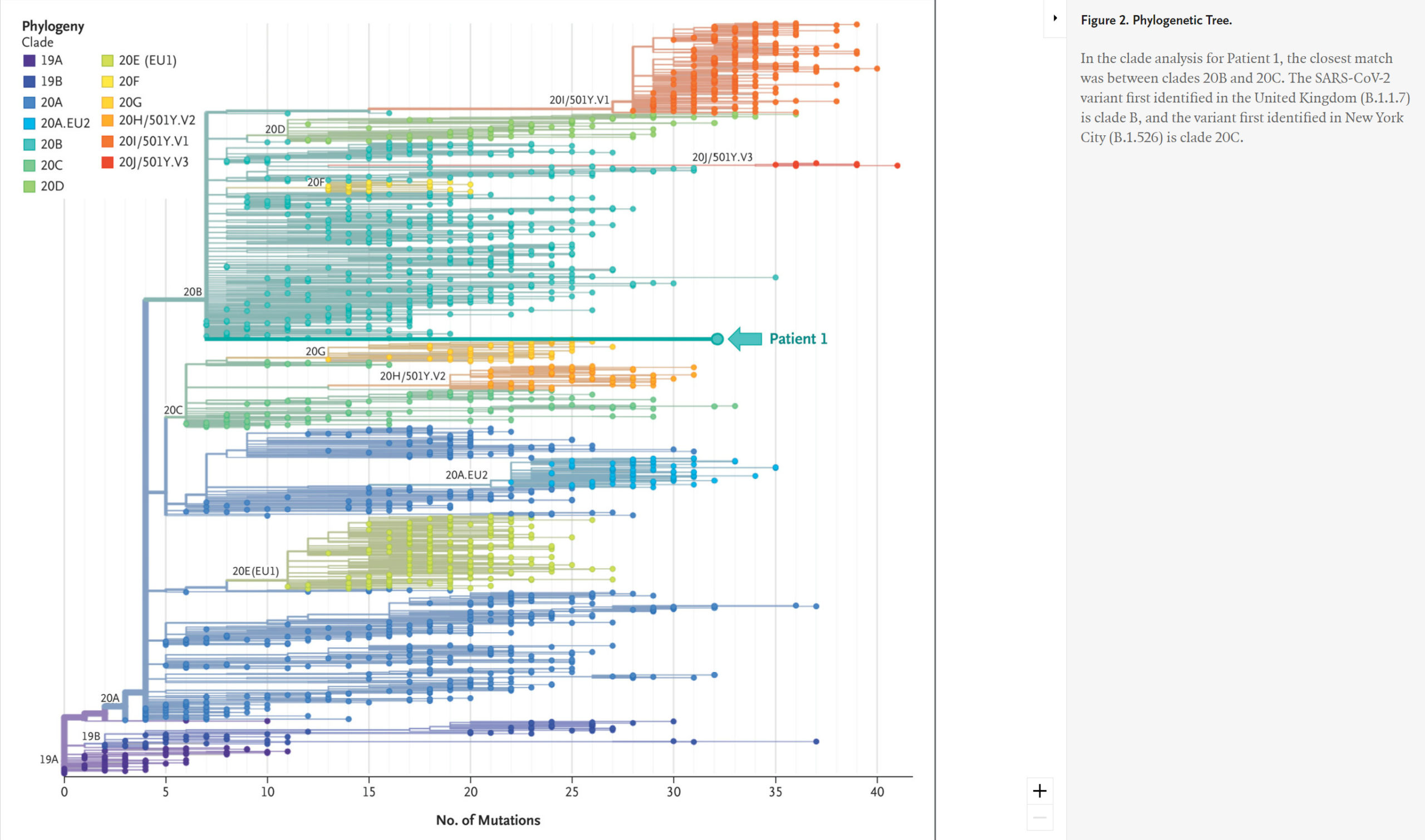
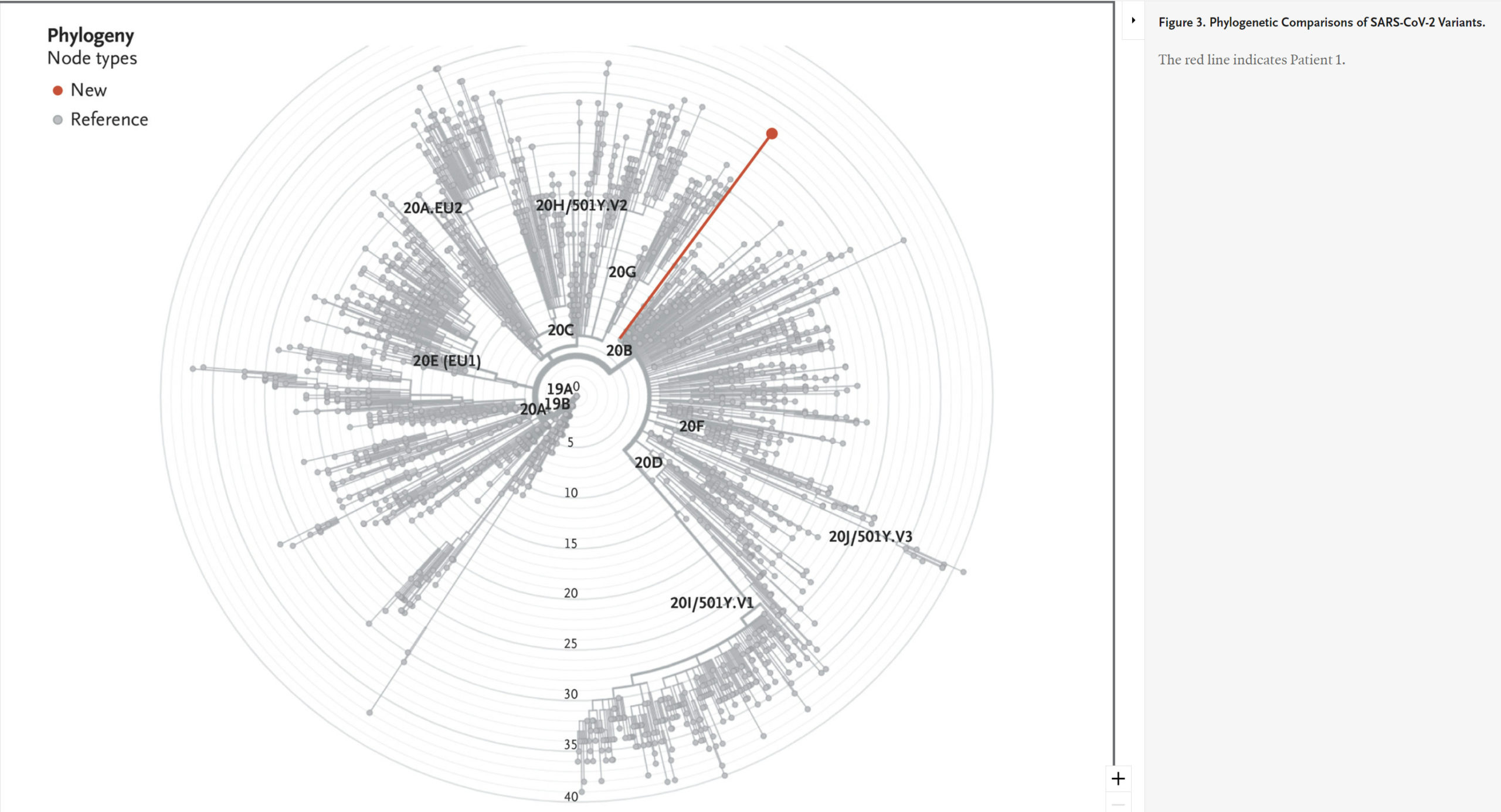
Phylogenetic Comparisons of SARS-CoV-2 Variants.
To explore the variants in Patient 1 in more detail, we used excess RNA available from extracted saliva to undertake whole viral genome sequencing. This analysis confirmed the sequence changes found in the S gene by targeted amplification and suggested that the infection resulted from a SARS-CoV-2 variant that is related to but distinct from the known variants of concern (the B.1.1.7 variant first identified in the United Kingdom and the B.1.526 variant first identified in New York City) (Figure 2 and Figure 3).

Neutralization Assays for Antibodies to the Wuhan-Hu-1 Isolate, the E484K Variant, and the B.1.526 Variant.
We therefore further tested the serum sample obtained from Patient 1 to measure its effectiveness against the wild-type virus, the E484K mutant, and the B.1.526 variant, and we found that the serum was equally effective against each (Figure 4). These data suggest that the antibody response in Patient 1 recognized these variants but was nonetheless insufficient to prevent a breakthrough infection.
Discussion
Clinical symptoms of Covid-19 developed 19 days after Patient 1 received the second dose of vaccine and 36 days after Patient 2 received the second dose. Both patients had histories consistent with a clinical response to vaccine boost. In Patient 1, documented high titers of neutralizing antibodies were present shortly after the development of symptoms. Although a baseline antibody test before illness and after vaccination would have been ideal, it remains possible that she became infected before the booster shot took full effect. Considering the clinical history, time course, and neutralizing antibody titers measured, we conclude that it is very likely that both patients had effective immune responses to the vaccines. Although these patients presented with clinically mild symptoms, it will be very important to ascertain whether severe symptoms can or cannot develop in others despite vaccination as variants continue to evolve.10 Taken together, our observations support the conclusion that we have characterized bona fide examples of vaccine breakthrough manifesting as clinical symptoms. Moreover, data from Patient 1 indicate that infection with variant virus can be sustained with a high viral load despite high levels of neutralizing antibodies to variants.
Examination of the SARS-CoV-2 sequences revealed that both patients were infected with variant viruses. Rapid identification of sequence variants by targeted PCR amplification showed that neither sequence precisely fit any known clade. Some of the substitutions in Patient 1 (T95I, del144, E484K, A570D, D614G, P681H, and D796H) were shared with B.1.526 (T95I, E484K, and D614G6), and three substitutions were shared with Patient 2 (in whom the variants T95I, G142V and del144, F220I, R190T, R237K, R246T, and D614G were detected). Whole viral genome sequencing revealed several additional substitutions, including D796H, present in a guanine–cytosine–rich region not identified by targeted PCR. These substitutions may decrease sensitivity to convalescent serum11 and may have some unique noncoding changes as compared with the clades first identified in Wuhan, the United Kingdom, and New York City.
Although more detailed analysis of whole-genome sequencing from Patient 1 was undertaken, we could not conclude that the variant in this patient was a Pango lineage because it was only present in a single person.12 Its closest links on the phylogenetic tree were the variant first identified in the United Kingdom (B.1.1.7) and the variant first identified in New York City (B.1.526), but with considerable differences (Figure 2 and Figure 3). It will be of interest to determine whether this may have resulted from a recombination event between B.1.1.7 and B.1.526, as has been recently reported for recombination between the B.1.1.7 lineage and the “wild-type” lineage first identified in Wuhan.13 Alternatively, shared substitutions may be the result of convergent evolution.
These observations in no way undermine the importance of the urgent efforts being taken at the federal and state levels to vaccinate the U.S. population. They also lend support to efforts to advance a new vaccine booster (as well as a pan-coronavirus vaccine) to provide increased protection against variants. In January 2021, Moderna announced clinical efforts to target a new variant of SARS-CoV-2 that was first identified in South Africa and includes three mutations (E484K, N501Y, and K417N) in the angiotensin-converting–enzyme 2 receptor-binding domain. These efforts are of critical value because recent studies have shown that immunizations are proving to be less potent against the variant first identified in South Africa (B.1.351), which might have acquired a partial resistance to neutralizing antibodies generated by natural infections or vaccinations.14,15 At the same time, our observations underscore the importance of the ongoing race between immunization and the natural selection of potential viral escape mutants. During this critical period, our data support the need to maintain layers of mitigation strategies, including serial testing of asymptomatic persons, open publication and analysis of vaccination and infection databases (such as those accruing data in New York City), and rapid sequencing of SARS-CoV-2 RNA obtained from a variety of high-risk persons.
Supplementary Material
| Supplementary Appendix | 86KB | |
| Disclosure Forms | 356KB |
Citing Articles (6)
-
(2021) Vaccine Breakthrough Infections with SARS-CoV-2 Variants. N Engl J Med DOI: 10.1056/NEJMc2107808.
-
Shekhar Kunal, Aditi, Kashish Gupta, Pranav Ish. (2021) COVID-19 variants in India : potential role in second wave and impact on vaccination. Heart & Lung 397.
-
Zulvikar Syambani Ulhaq, Gita Vita Soraya, Kristin Indriana. (2021) Breakthrough COVID-19 case after full-dose administration of CoronaVac vaccine. Indian Journal of Medical Microbiology 373.
-
Sarah N. Redmond, Lucas D. Jones, Navid Sadri, Christine Schmotzer, Maria E. Navas, Trina F. Zabarsky, Davinder Bhullar, Curtis J. Donskey. (2021) Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in vaccinated and unvaccinated healthcare personnel in a Veterans’ Affairs healthcare system. Infection Control & Hospital Epidemiology, 1-2.
-
Jasdeep Singh, Syed Asad Rahman, Nasreen Z. Ehtesham, Subhash Hira, Seyed E. Hasnain. (2021) SARS-CoV-2 variants of concern are emerging in India. Nature Medicine 13.
-
Kanika Tyagi, Amerta Ghosh, Dipti Nair, Koel Dutta, Prakash Singh Bhandari, Irshad Ahmed Ansari, Anoop Misra. (2021) Breakthrough COVID19 infections after vaccinations in healthcare and other workers in a chronic care medical facility in New Delhi, India. Diabetes & Metabolic Syndrome: Clinical Research & Reviews 15:3, 1007-1008.